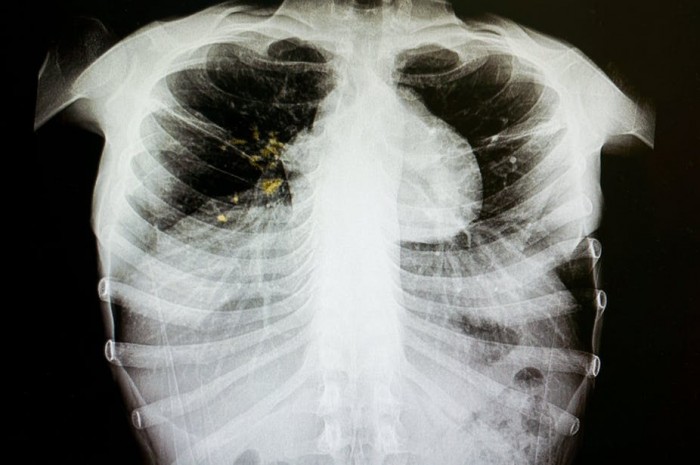
Tuberculosis (TB) is one of the leading causes of death from infectious diseases. Globally, it accounts for around 1.3 million deaths and 10.4 million people develop the disease every year.
The standard first-line treatment regimen used for newly diagnosed TB patients involves taking four different drugs over a period of six months. Some patients experience varying degrees of adverse reactions as a result. These factors have led to patients defaulting on their treatment. This, in turn, has led to a rise in drug resistance.
TB drug resistance – when the bacteria become resistant to at least one anti-TB drug – is a growing problem across the world. Current treatments that include more than one drug known as combination therapy have become inadequate because the bacteria have devised ways to survive even when antibiotics are used. An estimated 3.5% of new cases and 18% of previously treated cases of TB are drug resistant. There is therefore still a need to identify alternative drugs. But the drug discovery process is fairly lengthy (10 to 15 years). This timeline can be shortened with the aid of computers.
In an effort to find new ways of overcoming the problem, my colleagues and I at the South African National Bioinformatics Institute and school of pharmacy at the University of the Western Cape used a number of computer strategies to identify alternative drugs. The results were published recently in the Plos One Journal.
Our research presents a cost effective alternative way to tackle drug resistance by identifying drugs that prevent the bacteria from overcoming drug pressure. Using computer modelling, we were able to identify new drugs by building a reliable replica of the bacterial protein and then working out what the best orientation is for the drug to fit in the bacterial protein.
Ongoing studies using a combination of drugs will examine the effect of the drugs on stopping the growth of the bacterium. This, in turn, could lead to a new drug being included in the treatment regimen to beat drug-resistant TB.
Finding a solution
One way the bacteria is able to survive drug pressure, is by making proteins which mitigate the effect of the drug such as pumping the drugs out of the cell. Because some antibiotics are only effective when they are inside the bacterial cell, they become ineffective treatments.
The proteins that play this role are called efflux pumps. They’re very important targets in the drug discovery process. But it has been a struggle to identify drugs that can withstand the effect of the efflux pumps, with a number being dismissed as either too toxic or simply ineffective.
Using computer modelling we aimed to identify a drug that binds to the efflux pump protein.
Previously, the gold standard for identification of drugs with activity against the bacterium has been the use of wet-lab screening – growing the bug in a petri dish and adding drugs to it and measuring the loss of colony forming units. This is a high throughput method whereby large number of drugs can be added to different wells on a plate containing the growing bug. This approach is only feasible to specialised laboratories that not only fulfils biosafety standards but have large funding.
The approach has another drawback: many of the active drugs identified may bind to human drug targets that in turn could be harmful to the human host.
This is where the use of computers is proving useful. They can develop drugs that specifically target efflux pumps and reduce the harm to people. These computer-based methods have been used in the past to develop various drugs against several diseases like HIV, bacterial infections and Alzheimer’s disease.
Computers can simulate the work in a laboratory by performing complex calculations. This is powerful when working with thousands of small drug molecules that must be evaluated as a possible treatment option.
The first step in our research involved building a three dimensional shape of the protein using computer programmes. The next step was to fit the drugs into the protein shape and to find the best fit. This elimination process can take anything from a few weeks – if only a small number of computers are used – to a few days if a cluster of high-performance computers is used.
We used 24 computers which made the process faster and more efficient.
Cost benefit
Using a large number of computers for screening drugs reduces the cost and time spent testing drug candidates in laboratories. Another benefit is that it can eliminate drugs that are very likely to fail later in clinical trials.
The drugs identified by computer methods are very likely to lead to new and better treatments for drug-resistant TB, and will help to restore the action of some drugs being used in first-line treatment, such as Rifampicin, to which resistance has developed.
![]()
Ruben Cloete, Lecturer in Bioinformatics, University of the Western Cape
This article is republished from The Conversation under a Creative Commons license. Read the original article.